This project applies Support Vector Machines (SVM) to the Heart Disease dataset for the classification of patients into two categories: “No Disease” and “Disease.” The goal is to develop a robust predictive model that can assist healthcare professionals in identifying heart disease cases based on clinical and demographic variables.
For a detailed walkthrough of the analysis, refer to the complete project report:
📄 Heart Disease Diagnosis Report
.
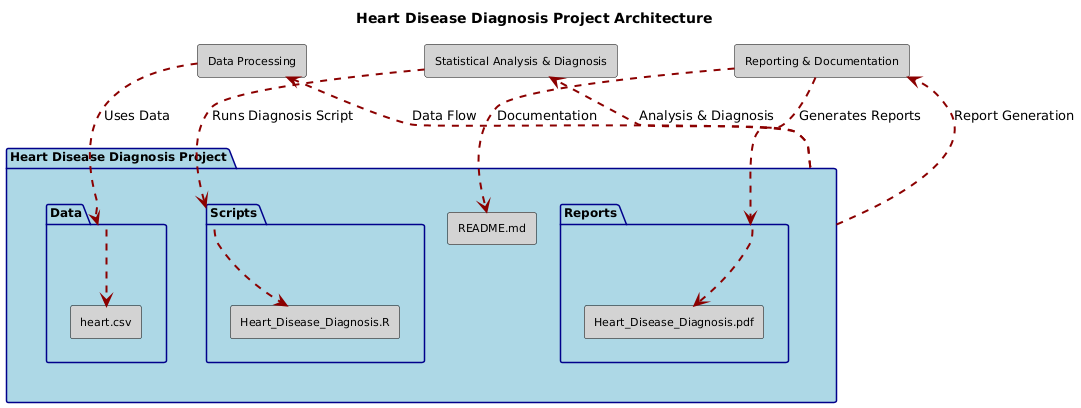
├── Data/
│ ├── heart.csv
├── Scripts/
│ ├── Heart_Disease_Diagnosis.R
├── Reports/
│ ├── Heart_Disease_Diagnosis.pdf
├── README.md
Feel free to reach out for feedback, questions, or collaboration opportunities:
LinkedIn: Dr. Syed Faizan
Author: Syed Faizan
Master’s Student in Data Analytics and Machine Learning
| #---------------------------------------------------------# | |
| # Syed Faizan # | |
| # Heart Disease Diagnosis using SVM's # | |
| # # | |
| # # | |
| # # | |
| # # | |
| #---------------------------------------------------------# | |
| #Starting with a clean environment---- | |
| rm(list=ls()) | |
| # Clearing the Console | |
| cat("\014") # Clears the console | |
| # Clearing scientific notation | |
| options(scipen = 999) | |
| #Loading the packages utilized for Data cleaning and Data Analysis----- | |
| library(tidyverse) | |
| library(grid) | |
| library(gridExtra) | |
| library(dplyr) | |
| library(kableExtra) | |
| library(ggplot2) | |
| library(DataExplorer) | |
| library(dlookr) | |
| library(lubridate) | |
| library(table1) | |
| library(psych) | |
| # Loading the initial Data set | |
| heartdf <- read.csv('heart.csv') | |
| str(heartdf) # confirming structure of the data set | |
| summary(heartdf) | |
| head(heartdf) | |
| tail(heartdf) | |
| any(is.na(heartdf)) # Analyzing and visualizing the missing data | |
| sum(is.na(heartdf)) | |
| plot_missing(heartdf) | |
| # Feature Engineering | |
| # Data transformation of target variable to factor | |
| heartdf$target <- factor(heartdf$target, labels = c("NoDisease", "Disease")) | |
| # Defining the label for each variable for clarity in the table | |
| label(heartdf$age) <- "Age" | |
| label(heartdf$sex) <- "Sex" | |
| label(heartdf$cp) <- "Chest Pain Type (CP)" | |
| label(heartdf$trestbps) <- "Resting Blood Pressure (trestbps)" | |
| label(heartdf$chol) <- "Cholesterol (chol)" | |
| label(heartdf$fbs) <- "Fasting Blood Sugar (fbs)" | |
| label(heartdf$restecg) <- "Resting ECG (restecg)" | |
| label(heartdf$thalach) <- "Maximum Heart Rate Achieved (thalach)" | |
| label(heartdf$exang) <- "Exercise-Induced Angina (exang)" | |
| label(heartdf$oldpeak) <- "ST Depression (oldpeak)" | |
| label(heartdf$slope) <- "Slope of ST Segment (slope)" | |
| label(heartdf$ca) <- "Number of Major Vessels (ca)" | |
| label(heartdf$thal) <- "Thalassemia (thal)" | |
| # Convert 'sex' to a factor with appropriate labels | |
| heartdf$sex <- factor(heartdf$sex, levels = c(0, 1), labels = c("Female", "Male")) | |
| # Convert 'cp' (chest pain type) to a factor with appropriate labels | |
| heartdf$cp <- factor(heartdf$cp, levels = c(0, 1, 2, 3), | |
| labels = c("Typical Angina", "Atypical Angina", "Non-Anginal Pain", "Asymptomatic")) | |
| # Convert 'fbs' (fasting blood sugar > 120 mg/dl) to a factor with appropriate labels | |
| heartdf$fbs <- factor(heartdf$fbs, levels = c(0, 1), labels = c("False", "True")) | |
| # Convert 'restecg' to a factor with appropriate labels | |
| heartdf$restecg <- factor(heartdf$restecg, levels = c(0, 1, 2), | |
| labels = c("Normal", "ST-T Wave Abnormality", | |
| "Left Ventricular Hypertrophy")) | |
| # Convert 'exang' (exercise-induced angina) to a factor with appropriate labels | |
| heartdf$exang <- factor(heartdf$exang, levels = c(0, 1), labels = c("No", "Yes")) | |
| # Convert 'slope' to a factor with appropriate labels | |
| heartdf$slope <- factor(heartdf$slope, levels = c(0, 1, 2), | |
| labels = c("Upsloping", "Flat", "Downsloping")) | |
| # Convert 'thal' to a factor with appropriate labels | |
| heartdf$thal <- factor(heartdf$thal, levels = c(0, 1, 2, 3), | |
| labels = c("Unknown", "Normal", "Fixed Defect", "Reversible Defect")) | |
| # Create table1 stratified by the target variable | |
| # Load necessary library | |
| library(table1) | |
| # Categorical variables | |
| table1(~ sex + cp + fbs + restecg + exang + slope + thal | target, | |
| data = heartdf) | |
| # Numerical variables | |
| table1(~ age + trestbps + chol + thalach + oldpeak + ca | target, | |
| data = heartdf) | |
| # Data Visualization | |
| # Create a data frame for categorical variables | |
| cat_df <- heartdf[, c("sex", "cp", "fbs", "restecg", "exang", "slope", "thal", "target")] | |
| # Create a data frame for numerical variables | |
| num_df <- heartdf[, c("age", "trestbps", "chol", "thalach", "oldpeak", "ca")] | |
| # Check the structure of the new data frames | |
| str(cat_df) | |
| str(num_df) | |
| # Checking if the numerical variables need scaling | |
| means <- sapply(num_df[, -7], mean) | |
| stddevs <- sapply(num_df[, -7], sd) | |
| scaling_check <- cbind(means, stddevs) | |
| print(scaling_check) | |
| # Scaling is needed, however it shall be done after visualization | |
| library(dlookr) | |
| library(DataExplorer) | |
| plot_outlier(num_df, col = "red") | |
| plot_normality(num_df, col = "yellow") | |
| plot_bar_category(cat_df) | |
| # Correlation Matrix | |
| library(ggcorrplot) | |
| ggcorrplot(cor(num_df), lab = TRUE) | |
| # Pair Plots | |
| library(GGally) | |
| ggpairs(num_df) | |
| # Scaling | |
| # Ensure the 'target' column is not scaled and is retained as is | |
| num_df <- num_df[, 1:6] # Extract the target column | |
| # Scale the remaining numerical columns (exclude 'target' column) | |
| num_df_scaled <- scale(num_df, center = TRUE, scale = TRUE) | |
| # Combine the scaled data with the 'target' column | |
| num_df <- data.frame(num_df_scaled) | |
| # Ensure 'target' is a factor in num_df | |
| # Convert 'target' in num_df to a factor with levels "NoDisease" and "Disease" | |
| num_df$target <- factor(heartdf$target) | |
| # Divide into test and train | |
| # Set seed for reproducibility | |
| set.seed(314) | |
| # Split the data into 70% training and 30% testing | |
| train_index <- sample(1:nrow(num_df_scaled), size = 0.7 * nrow(num_df_scaled)) | |
| # Create training and testing datasets | |
| train_data <- num_df[train_index, ] | |
| test_data <- num_df[-train_index, ] | |
| # Check dimensions of the train and test sets | |
| dim(train_data) # Should be 70% of the total rows | |
| dim(test_data) # Should be 30% of the total rows | |
| # SVM Implementation | |
| library(e1071) | |
| library(caret) | |
| # Train an SVM model with linear kernel on the training data | |
| svmfit <- svm(target ~ ., data = train_data, kernel = "linear", cost = 10, scale = FALSE) | |
| summary(svmfit) | |
| # Predictions on test data | |
| ypred <- predict(svmfit, newdata = test_data) | |
| # Confusion matrix using confusionMatrix from caret | |
| conf_matrix <- confusionMatrix(ypred, test_data$target) | |
| print(conf_matrix) | |
| # Tune SVM model to find best cost parameter | |
| set.seed(1) | |
| tune.out <- tune(svm, target ~ ., data = train_data, kernel = "linear", | |
| ranges = list(cost = c(0.001, 0.01, 0.1, 1, 10, 100))) | |
| summary(tune.out) | |
| # Use the best model found through tuning | |
| bestmod <- tune.out$best.model | |
| summary(bestmod) | |
| # Predictions using the best model on test data | |
| ypred_best <- predict(bestmod, newdata = test_data) | |
| # Confusion matrix for the best tuned model | |
| conf_matrix_best <- confusionMatrix(ypred_best, test_data$target) | |
| print(conf_matrix_best) | |
| # Support Vector Machine with Radial Kernel | |
| svmfit_radial <- svm(target ~ ., data = train_data, kernel = "radial", gamma = 1, cost = 1) | |
| summary(svmfit_radial) | |
| # Predictions using Radial Kernel SVM | |
| ypred_radial <- predict(svmfit_radial, newdata = test_data) | |
| # Confusion matrix for Radial Kernel | |
| conf_matrix_radial <- confusionMatrix(ypred_radial, test_data$target) | |
| print(conf_matrix_radial) | |
| # Tune SVM with Radial Kernel to find the best cost and gamma parameters | |
| set.seed(144) | |
| tune.out_radial <- tune(svm, target ~ ., data = train_data, kernel = "radial", | |
| ranges = list(cost = c(0.1, 1, 10, 100, 1000), gamma = c(0.5, 1, 2, 3, 4))) | |
| summary(tune.out_radial) | |
| # Use the best radial model found through tuning | |
| bestmod_radial <- tune.out_radial$best.model | |
| # Predictions using the best radial model | |
| ypred_best_radial <- predict(bestmod_radial, newdata = test_data) | |
| # Confusion matrix for the best radial model | |
| conf_matrix_best_radial <- confusionMatrix(ypred_best_radial, test_data$target) | |
| print(conf_matrix_best_radial) | |
| # ROC Curve for the Radial SVM Model | |
| library(ROCR) | |
| rocplot <- function(pred, truth, ...) { | |
| predob <- prediction(pred, truth) | |
| perf <- performance(predob, "tpr", "fpr") | |
| plot(perf, ...) | |
| } | |
| # Obtain decision values for plotting ROC | |
| svmfit_opt <- svm(target ~ ., data = train_data, kernel = "radial", gamma = 2, cost = 1, decision.values = TRUE) | |
| fitted <- attributes(predict(svmfit_opt, train_data, decision.values = TRUE))$decision.values | |
| # Plot ROC Curve for training data | |
| par(mfrow = c(1, 2)) | |
| rocplot(fitted, train_data$target, main = "Training Data (Radial Kernel)") | |
| # Obtain decision values for the test set | |
| fitted_test <- attributes(predict(svmfit_opt, test_data, decision.values = TRUE))$decision.values | |
| # Plot ROC Curve for test data | |
| rocplot(fitted_test, test_data$target, main = "Test Data (Radial Kernel)") | |
| # The Project Ends |

© 2025 Syed Faizan. All Rights Reserved.