This repository presents a 3D Liver and Liver Tumor Segmentation project using a UNet-based architecture. The model has been designed and trained to identify and segment liver and liver tumors from 3D medical images. The primary focus of this project was on model training, loss function analysis, and visual evaluation of the segmentation outputs.
The segmentation model is based on a 3D UNet architecture, optimized for volumetric medical image segmentation. The network features:
The above architecture helps the model efficiently learn the spatial hierarchies required for precise segmentation of liver and tumor regions.
The model was trained over 5 epochs, with training and validation loss tracked meticulously. Here’s the loss function plot that illustrates the convergence during the training process:
The steady decline in both training and validation loss demonstrates the effectiveness of the UNet model in learning the complex patterns in liver segmentation.
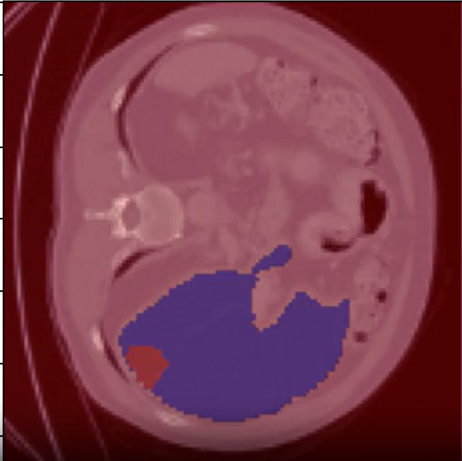
A qualitative assessment was conducted using a single test image to evaluate the segmentation performance visually. The model’s output shows clear demarcation of the liver and tumor regions, compared to the ground truth mask.
The overlay highlights:
(128, 128, 128).# Clone the repository
git clone https://github.com/SYEDFAIZAN1987/3D-Liver-and-Liver-Tumor-Segmentation.git
# Navigate to the project folder
cd 3D-Liver-and-Liver-Tumor-Segmentation
# Install dependencies
pip install -r requirements.txt# Training the model
python 3d_liver_and_liver_tumor_segmentation_.py
# Visualizing results
# The output snapshots and loss plots will be saved in the results folderThis project is licensed under the MIT License.
Developed by Syed Faizan.

© 2025 Syed Faizan. All Rights Reserved.